Lysosomes signal through the epigenome to regulate longevity across generations
(how to read a paper and extract summary from the paper to learn)
by Ellie S.
Agenda
- Read the Abstract & provide background
- Reading other parts of the paper
- Results
- Conclusion
- Future Directions
What do we think they are doing?
“The epigenome is sensitive to metabolic inputs and is crucial for aging. Lysosomes act as a signaling hub to sense metabolic cues and regulate longevity. We found that lysosomal metabolic pathways signal through the epigenome to regulate transgenerational longevity in Caenorhabditis elegans. Activation of lysosomal lipid signaling and lysosomal adenosine monophosphate–activated protein kinase (AMPK) or reduction of lysosomal mechanistic target of rapamycin (mTOR) signaling increased the expression of a histone H3.3 variant and increased its methylation on K79, leading to life-span extension across multiple generations. This transgenerational prolongevity effect required intestine-togermline transportation of histone H3.3 and a germline-specific H3K79 methyltransferase and was recapitulated by overexpressing H3.3 or the H3K79 methyltransferase. Thus, signals from a lysosome affect the epigenome and link the soma and germ line to mediate transgenerational inheritance of longevity.”
Identify key words

How did they do it?
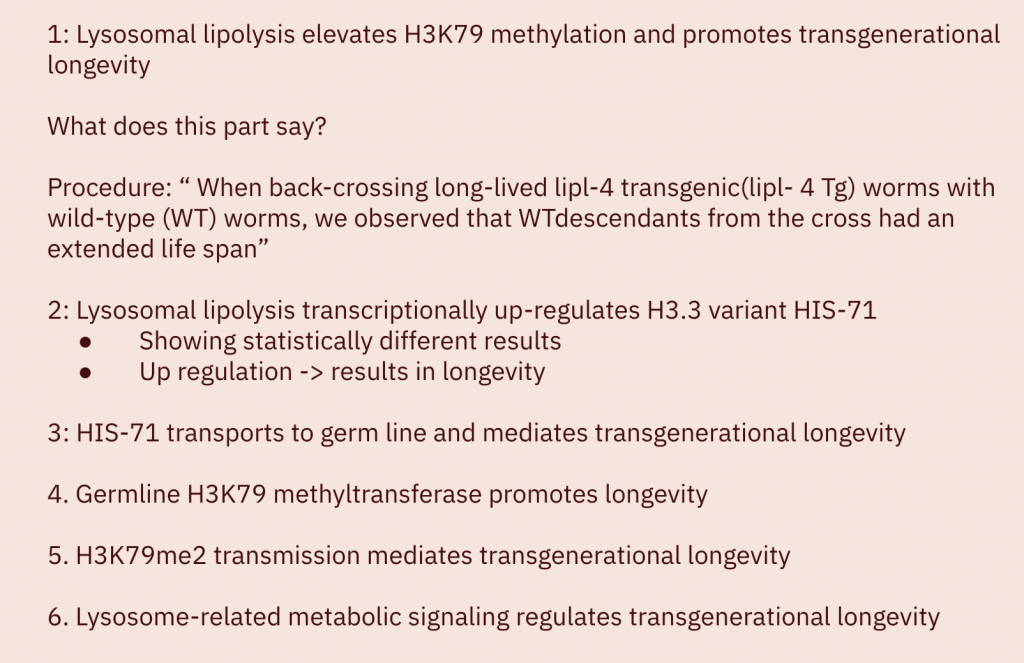
Conclusion / Discussion
“Based on our data, we propose the following model (Fig. 5R). Induced lysosomal lipolysis triggers lysosomal AMPK activation and mTORC1 suppression, up-regulating the transcription of H3.3, which is transported to the germ line, modified by H3K79 methyltransferase, and transmitted across generations to promote longevity. H3.3-enriched epigenomes exhibit distinct transcriptional responses from canonical H3–enriched ones (13, 14). The H3.3 to H3 transition restricts embryonic pluripotency during C. elegans development (20). H3.3 can help to maintain genome stability (32–34) and repair DNA damage (35, 36). Increased H3.3 incorporation may preserve a youthful epigenomic state during aging.
Our findings reveal cross-tissue epigenetic coordination in transgenerational longevity, mediated by the yolk-based transport of histones. In Drosophila, maternal histones are transported from nurse cells to oocytes with yolk lipids (37). Yolk proteins resemble mammalian very-low-density lipoproteins (38); however, whether lipoproteins facilitate histone trafficking in mammals remains unknown.”
Future Directions
Initiation: Induced lysosomal lipolysis occurs.
Signaling Cascade: Lysosomal lipolysis triggers lysosomal AMPK activation and mTORC1 suppression.
Transcriptional Activation: This signaling cascade leads to the up-regulation of the transcription of H3.3 (likely referring to the his−71 gene mentioned in the previous text).
Transport: The newly synthesized H3.3 is transported to the germ line (mediated by yolk-based transport, suggesting cross-tissue epigenetic coordination).
Epigenetic Modification: In the germ line, H3.3 is modified by H3K79 methyltransferase.
Inheritance and Outcome: The modified H3.3 is transmitted across generations to promote longevity.
References
Note: this is intended to give summary of the paper, there are direct conclusion and discussions from the paper. All data and writing belong to the original authors: https://www.science.org/doi/10.1126/science.adn8754






